Now to instigate Deep Learning. As opposed to the Random forest method, using a fully convoluted network such as Google's Inception does not require features to be extracted using custom coded algorithms targeted to the animal's echo location voice. The network works it all out on it's own and a good pre-trained model already has a shed load of features defined which can be recycled and applied to new images which are completely unrelated to the old ones. Sounds too good to be true?
To start with, I was very sceptical about it being able to tell the difference between identical calls at different frequencies, which is important when trying to classify members of the Pipistrelle genus. Basically, calls above 50 KHz are from Soprano Pips and calls under 50 KHz are Common Pips (There are other species in this genus, but not where I live!). So can the network tell the difference? The answer is both yes and no since we are forgetting one major tool at our disposal - data augmentation.
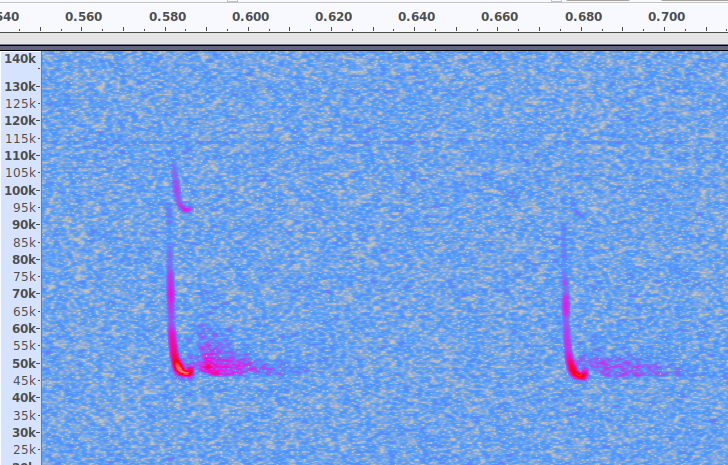
Data augmentation can take many different forms such as flipping the image either horizontally or vertically or both, to give us 4x more data. Great for photos of dogs, but totally inappropriate for bat calls ! (bats never speak in reverse or upside down). Scaling and cropping is also inappropriate as we need to keep the frequency axis in tact. Possibly the only thing we can do is move the main echo-location calls in the time axis ..... so if we have a 20 msec call in a 500 msec audio file we could shift that call sideways in the time frame as much as we wanted. I chose to shift it (about) 64 times with some simple code to create a 'sliding window'. The code uses the Bash 'Mogrify' command which, strangely, only works properly on .jpg images.
Essentially, it involves 8 of these:
# Convert all .png files to .jpg or else mogrify wont work properly:
ls -1 *.png | xargs -n 1 bash -c 'convert "$0" "${0%.png}.jpg"'
# delete the .png files:
find . -maxdepth 1 -type f -iname \*.png -delete
# Now split up all the 0.5 second long files into 8 parts of 680 pixels each:
for file in *
do
fname="${file%.*}"
mogrify -crop 5500x680+220+340 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname".jpg
# This produces image 1 of 8:
cp "$fname".jpg "$fname"_1.jpg
mogrify -crop 680x680+0+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_1.jpg
# This produces image 2 of 8:
cp "$fname".jpg "$fname"_2.jpg
mogrify -crop 680x680+680+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_2.jpg
# This produces image 3 of 8:
cp "$fname".jpg "$fname"_3.jpg
mogrify -crop 680x680+1360+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_3.jpg
# This produces image 4 of 8:
cp "$fname".jpg "$fname"_4.jpg
mogrify -crop 680x680+2040+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_4.jpg
# This produces image 5 of 8:
cp "$fname".jpg "$fname"_5.jpg
mogrify -crop 680x680+2720+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_5.jpg
# This produces image 6 of 8:
cp "$fname".jpg "$fname"_6.jpg
mogrify -crop 680x680+3400+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_6.jpg
# This produces image 7 of 8:
cp "$fname".jpg "$fname"_7.jpg
mogrify -crop 680x680+4080+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_7.jpg
# This produces image 8 of 8:
cp "$fname".jpg "$fname"_8.jpg
mogrify -crop 680x680+4760+0 /media/tegwyn/Xavier_SD/dog-breed-identification/build/plecotus_test_spectographs/test/"$fname"_8.jpg
i=$((i+1))
done
The final code is a bit more complicated than this, but not by much!
Suddenly we've got about 64x the amount of data. But what do the images contain? What if they contain the gaps between calls - ie nothing? .... So now the images needed to be inspected one at a time to make sure that they actually had relevant content .... All 30,000 of them! At a processing rate of about 2 per second, this took about 4 hours. That's 4 hours of sitting in front of a screen pressing the delete and forward arrow. Was it worth it?
I'm not going to go through the process of setting up the software environment for training the Inception classifier using Tensorflow on an Nvidia GPU as most probably, by the time I've finished typing it out, it will have changed. I used to be able to use my Jetson Xavier to train on Nvidia's DetectNet, but guess what? .... Yes, the software dependancies changed slightly and the system won't run without unfathomable critical errors.
Also, it's the age old thing, there's dozens of tutorials for classifying images and 95% of them are incomplete, irrelevant, out of date or simply dont work. After a lot of code skim reading a bit of trial and error, I settled on THIS ONE.
What's great is that the data is really well organised in a very simple manner and the code is well documented and easy to use. Only 2 main files: retrain.py and classify.py. It's supposed to classify dog breeds, but works perfectly on bat spectograms as well!
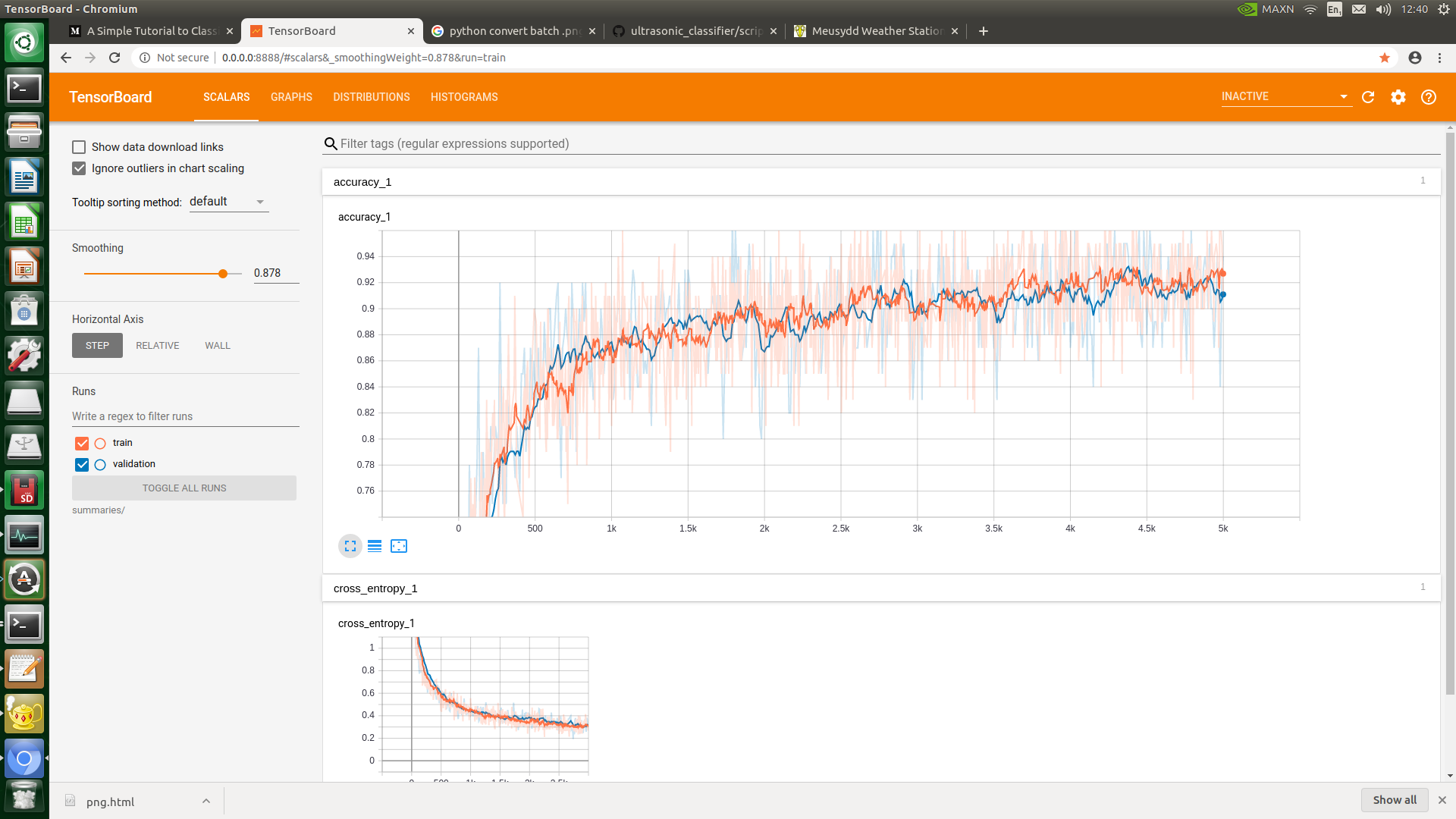
Throw all the species into their own folder, label the folder with the species, chuck the labelled folders into the 'dataset' folder, delete all the dog stuff and run the retrain.py script. Very simple. After training, find the 'retrained_labels.txt' and change the first few lines according to bat species after a few tests. Test the classifier on fresh data by putting it into the 'test' folder and run the classifier.py script. During training, even the Tensorboard function worked:
Now test a batch of spectograph images, preferably unseen by the training:
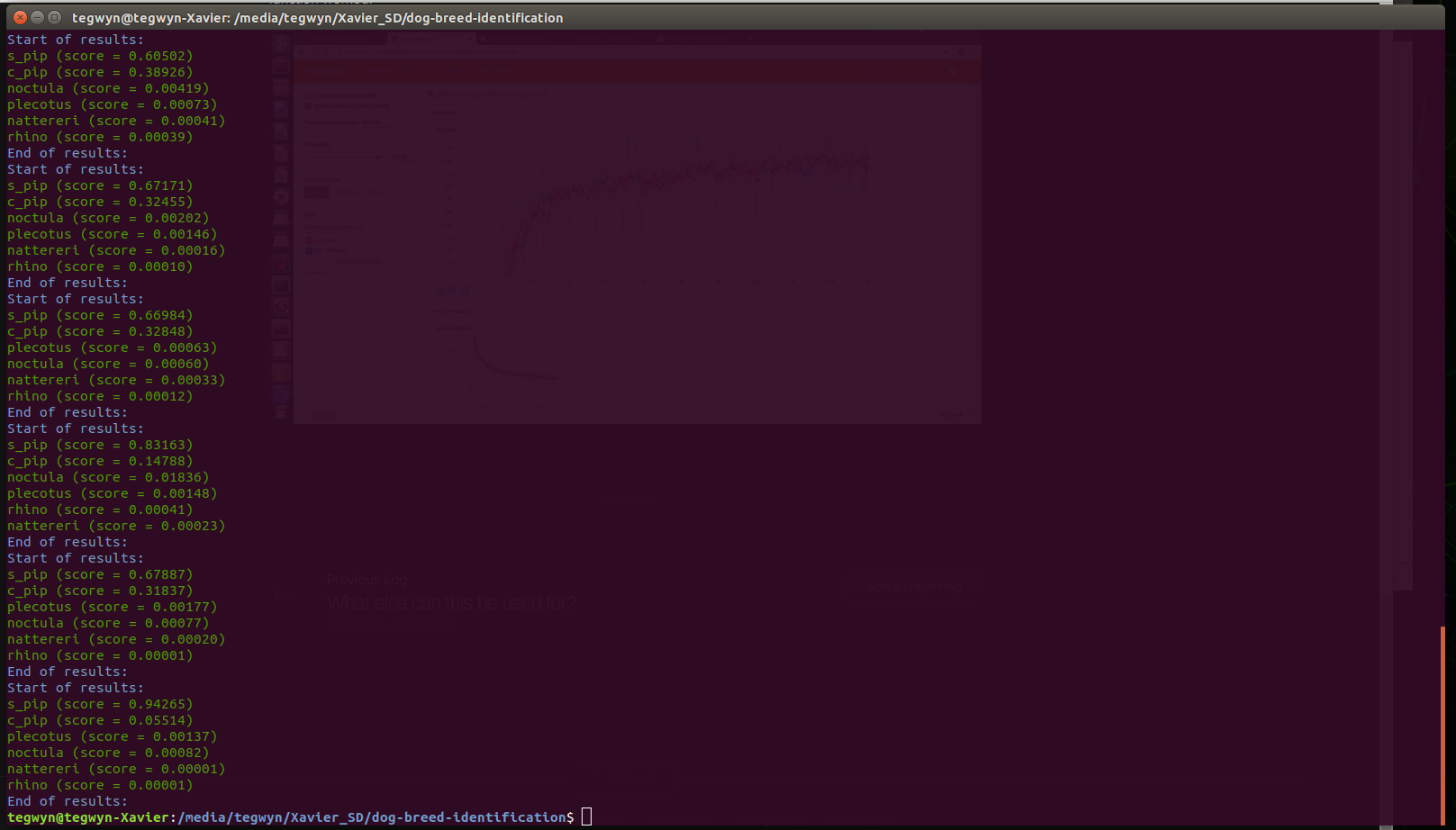
Fortunately, the classifier worked properly on the Pipistrelle species, correctly classifying all the Sopranos as sopranos. What's interesting is that common pips is a close second, and sometimes very close, which is exactly as we would expect. Great - the system is working!
 Capt. Flatus O'Flaherty ☠
Capt. Flatus O'Flaherty ☠
Discussions
Become a Hackaday.io Member
Create an account to leave a comment. Already have an account? Log In.